Tamil Iniyan Gunasekaran, et al. – Columbia University.
Background: While AD risk is known to be highly heritable, relatively few pathogenic mutations have been identified. Discovering new pathogenic mutations could enable screening for these mutations as well as investigation into how they contribute to AD pathology.
This Study: Gunasekaran and colleagues examined sequencing data from families with AD history located in the US and the Dominican Republic.
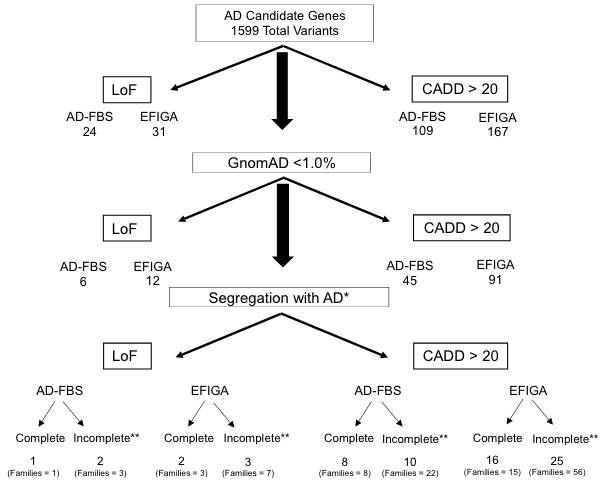
- Their search was restricted to mutations predicted to change the protein-coding sequence or induce early protein truncation in genes flagged by previous GWAS studies.
- 27 novel variants were identified in either cohort, with ABCA7 and AKAP9 contributing the highest number of variants. Only one variant, a rare loss-of-function mutation to ABCA7, was present in both cohorts.
- In many US families and most Dominican families, no mutations to the studied genes, nor APOE4, were present, underscoring the work needed to expand the pool of known risk genes.
Bottom Line: While analysis of sequencing data from families with AD risk uncovered new pathogenic mutations, these pathogenic mutations fail to account for much of AD risk.
Open Question: While the goal of this study led these researchers to only study mutations affecting protein-coding sequences, numerous other studies have highlighted the importance of mutations to regulatory sequences in and around these genes. What fraction of the families without pathogenic mutations carry variants to these regulatory regions?