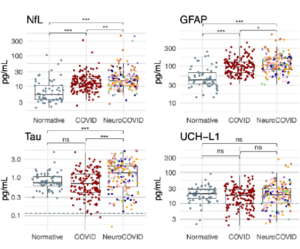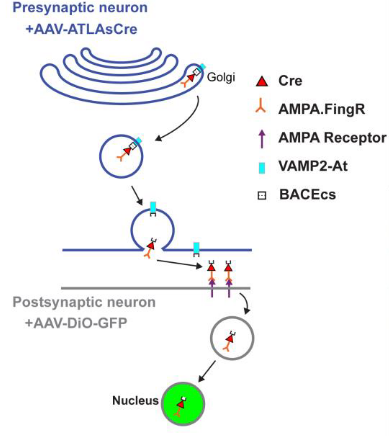
“ATLAS: A rationally designed anterograde transsynaptic tracer.”
Jacqueline F. Rivera, et al. – University of Southern California.
In this study, the Arnold Lab reports the generation of ATLAS, a monosynaptic anterograde tracer. Previous constructs from this group used virally-transduced intrabodies, which bind to proteins of choice, to label synaptic proteins. ATLAS fuses an intrabody against the glutamate receptor subunit GluA1 with peptides targeting the resulting protein for release via synaptic vesicles; the protein is endocytosed by the postsynaptic neuron following glutamate receptor binding. Variants that include Cre or FLP recombinase allow for labeling of postsynaptic cells in tracing experiments. This approach addresses major issues with existing viral anterograde tracers, such as undesired retrograde labeling and toxicity of many viral vectors.
The chief drawback of ATLAS is that, for unknown reasons, ATLAS-Cre does not efficiently and consistently label cells bearing a genomic reporter allele. Sufficient labeling only occurred when the reporter construct was delivered with a viral vector. Unbiased tracing experiments should still be viable with the use of AAV serotypes, such as PHP.eb, that can be administered systemically, expressing the reporter throughout the nervous system. Since the issues with other anterograde tracers have no effective mitigating solution, however, ATLAS is still positioned to answer the need for an anterograde transsynaptic tracer.